February digest highlights the current state of vaccines against resistant bacterial pathogens and novel ways by which they can evolve resistance, effects of common sweeteners on conjugation rate and how international travel contributes to spread of multidrug-resistant bacteria, and much more. Check out our selection of AMR focused podcast episodes and webinars. Happy reading!
Global
Newsletter: WHO Antimicrobial Resistance Newsletter 2021 – WHO
Review: Antimicrobial resistance and COVID-19: Intersections and implications – Gwenan M. Knight – eLife
Review: The role of vaccines in combatting antimicrobial resistance – Francesca Micoli – Nature Reviews Microbiology
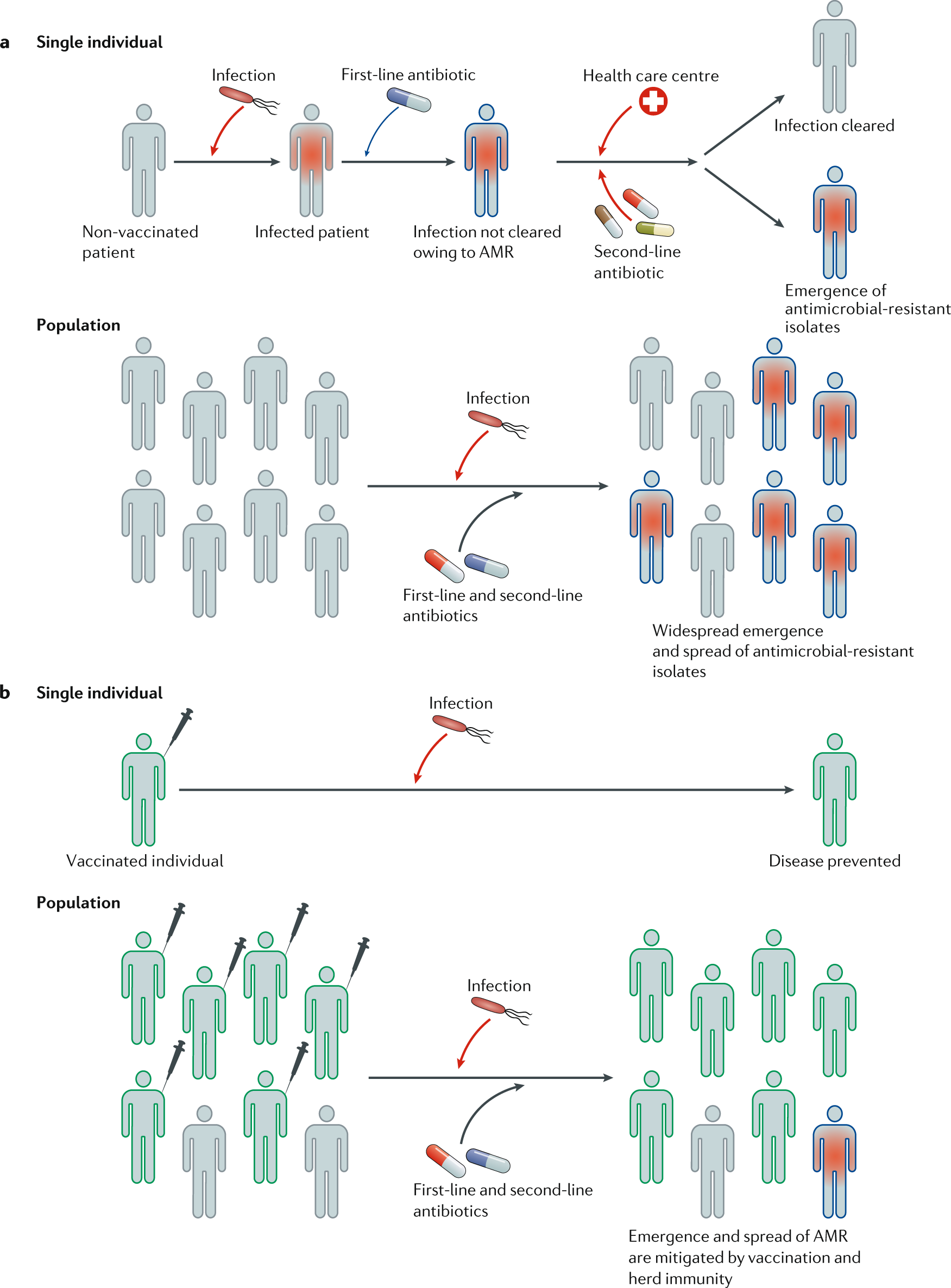
Review: Beyond Antimicrobial Use: A Framework for Prioritizing Antimicrobial Resistance Interventions – Noelle R. Noyes – Annual Review of Animal Biosciences
Clinically relevant mutations in core metabolic genes confer antibiotic resistance – Allison J. Lopatkin – Science
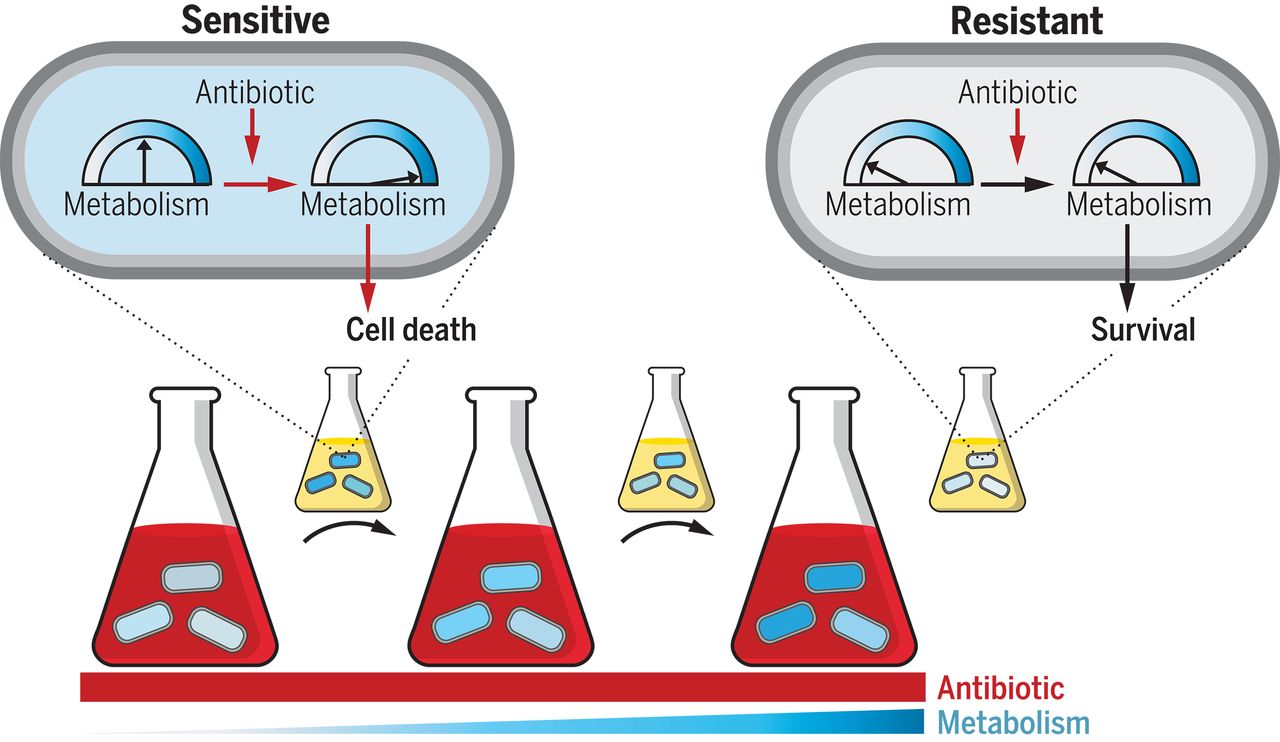 *Mutations targeting central carbon and energy metabolism arise in response to antibiotic treatment. These mutations confer resistance and are highly prevalent in clinical pathogens, expanding the known means by which pathogenic microbes can evolve resistance.
*Mutations targeting central carbon and energy metabolism arise in response to antibiotic treatment. These mutations confer resistance and are highly prevalent in clinical pathogens, expanding the known means by which pathogenic microbes can evolve resistance.
Nonnutritive sweeteners can promote the dissemination of antibiotic resistance through conjugative gene transfer – Zhigang Yu – ISME Journal
*Exposure to nonnutritive sweeteners (saccharine, sucralose, aspartame) enhances conjugation in bacteria.
Using ecological coexistence theory to understand antibiotic resistance and microbial competition – Andrew D. Letten – Nature Ecology & Evolution
Human resistome
Characterization of the human skin resistome and identification of two microbiota cutotypes – Zhiming Li – Microbiome
Dynamics of intestinal multidrug-resistant bacteria colonisation contracted by visitors to a high-endemic setting: a prospective, daily, real-time sampling study – Anu Kantele – Lancet Microbe
*International travel contributes substantially to the global spread of intestinal multidrug-resistant Gram-negative bacteria. Real-time sampling study of 20 European visitors to Laos volunteered to provide daily stool samples.
Quantifying the effects of antibiotic treatment on the extracellular polymer network of antimicrobial resistant and sensitive biofilms using multiple particle tracking – Lydia C. Powell – npj Biofilms Microbiomes
Soil
Bacterial communities regulate temporal variations of the antibiotic resistome in soil following manure amendment – Jianhua Cheng – Environmental Science and Pollution Research
Water environment
Microbiota analysis of rural and urban surface waters and sediments in Bangladesh identifies human waste as driver of antibiotic resistance – Ross Stuart McInnes – bioRxiv
Antibiotic Resistance in Wastewater Treatment Plants and Transmission Risks for Employees and Residents: The Concept of the AWARE Study – Laura Wengenroth – medRxiv
Animal studies
Metagenomics of antimicrobial and heavy metal resistance in the cecal microbiome of fattening pigs raised without antibiotics – Paiboon Tunsagool – Applied and Environmental Microbiology
Bioinformatics
A roadmap for the generation of benchmarking resources for antimicrobial resistance detection using next generation sequencing – Mauro Petrillo – F1000Research
HMD-ARG: hierarchical multi-task deep learning for annotating antibiotic resistance genes – Yu Li – Microbiome
Critical evaluation of short, long, and hybrid assembly for contextual analysis of antibiotic resistance genes in complex environmental metagenomes – Connor L. Brown – Scientific Reports
Critical evaluation of the impact of assembly leveraging short reads, nanopore MinION long-reads, and a combination of the two (hybrid) on ARG contextualization using seven prominent assemblers.
PathoFact: a pipeline for the prediction of virulence factors and antimicrobial resistance genes in metagenomic data – Laura de Nies – Microbiome
Techniques
Creation of Universal Primers Targeting Nonconserved, Horizontally Mobile Genes: Lessons and Considerations – Damon C. Brown and Raymond J. Turner – Applied and Environmental Microbiology
Robustness in quantifying the abundance of antimicrobial resistance genes in pooled faeces samples from batches of slaughter pigs using metagenomics analysis – Vibe Dalhoff Andersen – Journal of Global Antimicrobial Resistance
Webinars and conferences
Resistomap Webinar Series: Monitoring ARGs in LMICs – Resistomap – 30 March 2021
CAMDA 2021 Conference – ISMB ECCB – 26-27 July 2021
Data analysis contest challenges: The Metagenomic Phage Forensics of Anti-Microbial Resistance
The Initiative Seminar Engineering Health: Joining forces in healthcare solutions – Chalmers University of Technology in Gothenburg – 13-14 April 2021
*Johan Bengtsson-Palme will talk about Antibiotic resistance as a health and research challenge.
Learning from COVID-19 to tackle the silent pandemic of antibiotic resistance – GARDP – 4 March 2021
AMR Innovation Mission UK 2021 – AMR Insights – 10-12 May 2021
The One Health EJP Annual Scientific Meeting – One Health EJP – 9-11 June 2021
Podcast
The AMR studio: Ep 25. Linus Sandegren & mobile genetic elements. Identifying resistance origins. Reimbursing ABs. – Uppsala Antibiotic Center – 1 February 2021
Dr. Heer Mehta: Antibiotic Resistance and Experimental Evolution – The Bioinformatics and Beyond Podcast – 6 February 2021
Courses
Tackling Antibiotic Resistance: What Should Dental Teams Do? – British Society for Antimicrobial Chemotherapy and FDI World Dental Federation
Discover the danger posed by antibiotic resistance, and how dental teams can meet the challenge to protect patients.




 *Mutations targeting central carbon and energy metabolism arise in response to antibiotic treatment. These mutations confer resistance and are highly prevalent in clinical pathogens, expanding the known means by which pathogenic microbes can evolve resistance.
*Mutations targeting central carbon and energy metabolism arise in response to antibiotic treatment. These mutations confer resistance and are highly prevalent in clinical pathogens, expanding the known means by which pathogenic microbes can evolve resistance.