prepared by Ioannis Kampouris
This week, the @EMBARK_JPIAMR team is at the World Microbe Forum:
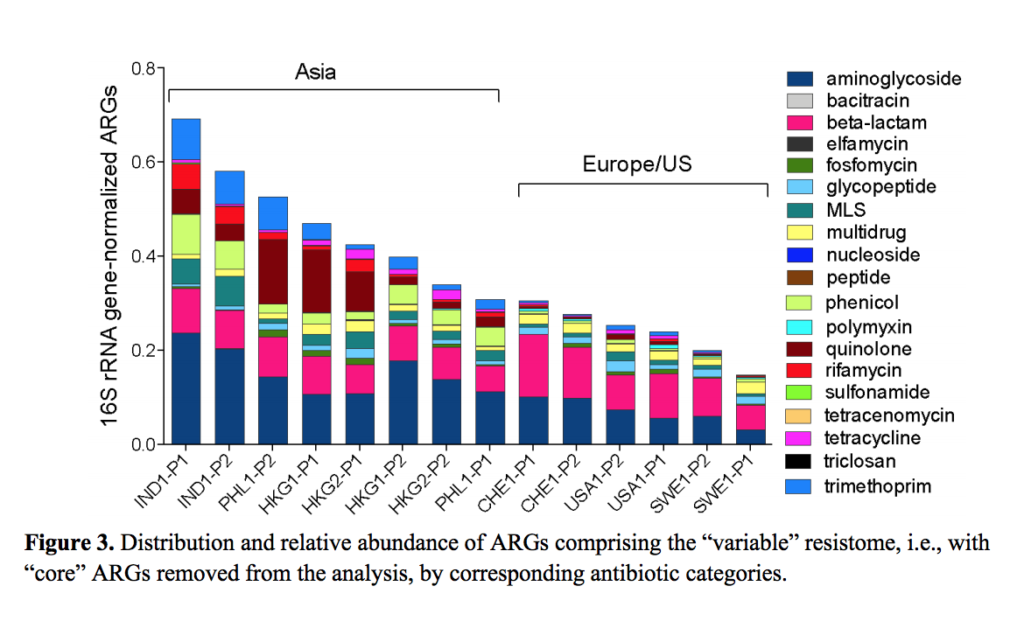
Svetlana Ugarcina Perovic and Luis Pedro Coelho with their work on ARGs at global scale,
Anna Abramova, Thomas Berendonk and Johan Bengtsson-Palme on ARGs across environments,
Saba Yasmin and Rabaab Zahra on E. coli in sewage water.
We would like to invite you for the 4th #EMBARK2021 webinar with Kimberly Kline and Sofia Forslund!
Global
There is no market for new antibiotics: this allows an open approach to research and development – Dana M. Klug, Fahima I. M. Idiris – Welcome Open Research
*This opinion paper suggests that there is a necessity for a new open source model for antibiotic discovery, which should be independent from whether a market for new antibiotics exist or not.
Destination shapes antibiotic resistance gene acquisitions, abundance increases, and diversity changes in Dutch travelers – Alaric W. D’Souza, Manish Boolchandani – Genome Medicine
*The authors claim that travel shapes the architecture of the human gut resistome and results in AMR gene acquisition against a variety of antimicrobial drug classes.
Soil environment
Modeling the vertical transport of antibiotic resistance genes in agricultural soils following manure application – Renys E. Barrios – Environmental Pollution
*The authors claim that detachment and decay coefficients for ARGs in soil were not affected by manure, while the load of ARGs in leachate was not significantly affected by manure.
Wastewater Treatment
Antibiotic transformation in an anaerobic membrane bioreactor linked to membrane biofilm microbial activity – Moustapha Harb – Environmental Research
Wastewater Based Epidemiology Enabled Surveillance of Antibiotic Resistance – M.V. Riquelme – MedRxiv
*The Wastewater Based Epidemiology approach helped to identify geographic hot-spots where attention towards curbing the spread of antibiotic resistance may be particularly warranted.
Hospital environment
A species-wide genetic atlas of antimicrobial resistance in Clostridioides difficile – Korakrit Imwattana – BioRxiv
*This review provides insights in the global C. difficile population, which may help in the early detection of drug-resistant C. difficile strains.
Human experiments
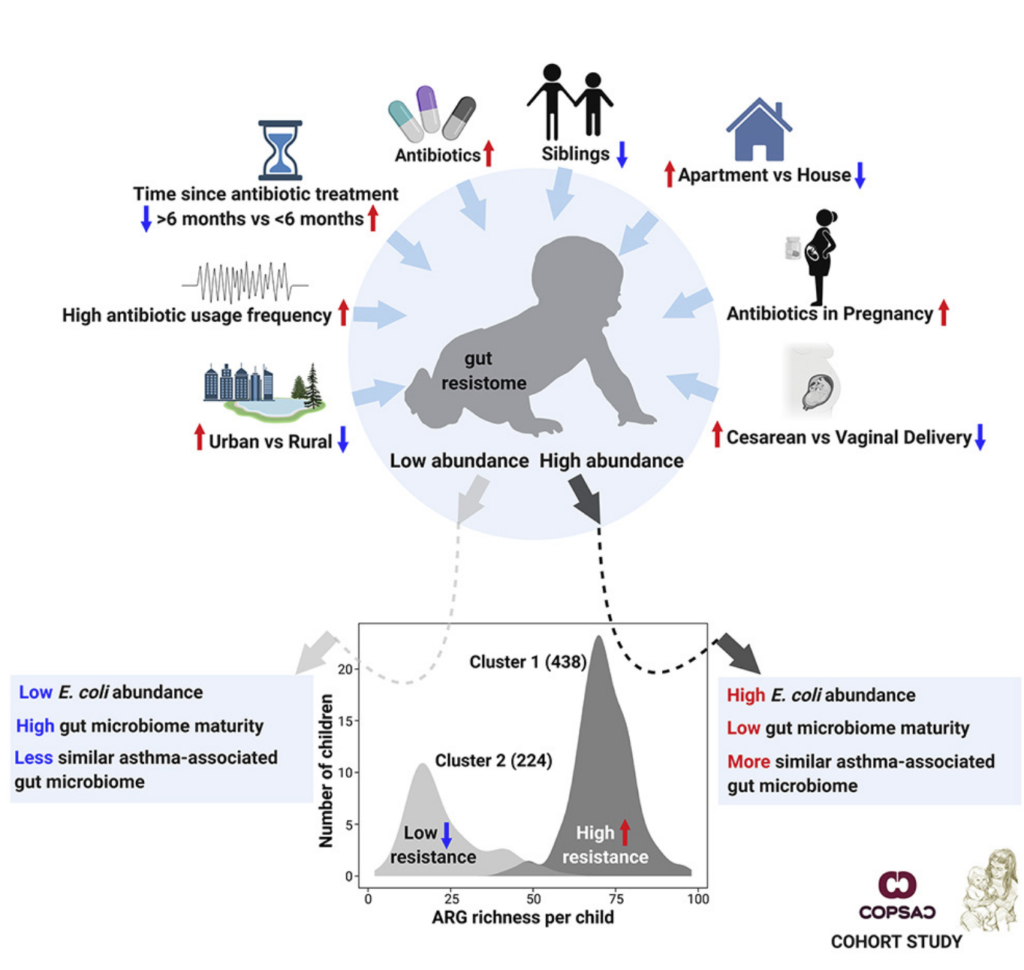
The infant gut resistome associates with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition – Xuanji Li – Cell Host & Microbiome
*The study reveals a clear bimodal distribution of ARG richness that is driven by the composition of the gut microbiome, especially E. coli.
Agricultural applications
Chitosan as additive affects the bacterial community, accelerates the removals of antibiotics and related resistance genes during chicken manure composting – Hongdou Liu – Science of Total Environment
*The use of chitosan can enhance the remove of antibiotics and antibiotic resistance genes during manure composting.
Animal experiments
Chironomidae larvae: A neglected enricher of antibiotic resistance genes in the food chain of freshwater environments – Chengshi Ding – Environmental Pollution
*Feeding experiments further confirmed that ARGs from Chironomidae larvae can be transferred to the fish gut.
Techniques
A Peek into the Plasmidome of Global Sewage – Philipp Kirstahler – mSystems
*The authors describe a laboratory and bioinformatics workflow for the recovery of plasmids and other potential extrachromosomal DNA elements from complex microbiomes.
Validation of selective agars for detection and quantification of Escherichia coli resistant to critically important antimicrobials – Zheng Z Lee – bioRxiv
Bioinformatics
Predicting Antimicrobial Resistance Using Partial Genome Alignments – D. Aytan-Aktug – mSystems
Review: Antibiotic resistance: Time of synthesis in a post-genomic age – Teresa Gil-Gil – Computational and Structural Biotechnology Journal
Horizontal Gene Transfer
Quantitative analysis of horizontal gene transfer in complex systems – Jenifer Moralez – Current Opinion in Microbiology
Phages
Comprehensive Scanning of Prophages in Lactobacillus: Distribution, Diversity, Antibiotic Resistance Genes, and Linkages with CRISPR-Cas Systems – Zhangming Pei – mSystems
*Uneven prophage distribution, with highly diverse genome features and distinct clusters based on host species along with detection of ARGs in Lactobacillus prophages.
L-form switching confers antibiotic, phage and stress tolerance in pathogenic Escherichia coli – Aleksandra Petrovic Fabijan – bioRxiv
Podcast
Superbugs and You – True Stories from Scientists and Patients around the World – The Antimicrobial Resistance Fighter Coalition
The @EMBARK_JPIAMR team wishes you happy Summer holidays!