March digest features review on nonlytic antimicrobial functions of complement, metabolic fitness models, BActeriophage SElection for your Laboratory (a collection of representative E. coli phages), Dr Dirt game, and more. Check out our list of AMR focused podcasts. Enjoy!
And, we are exciting to share with you EMBARK NEWS – the SPRING AMR WEBINARS! Join us and our invited speakers to discuss the latest AMR insights. Register for the first webinar on April 28 with Gerry Wright and Luis Pedro Coelho HERE.
General
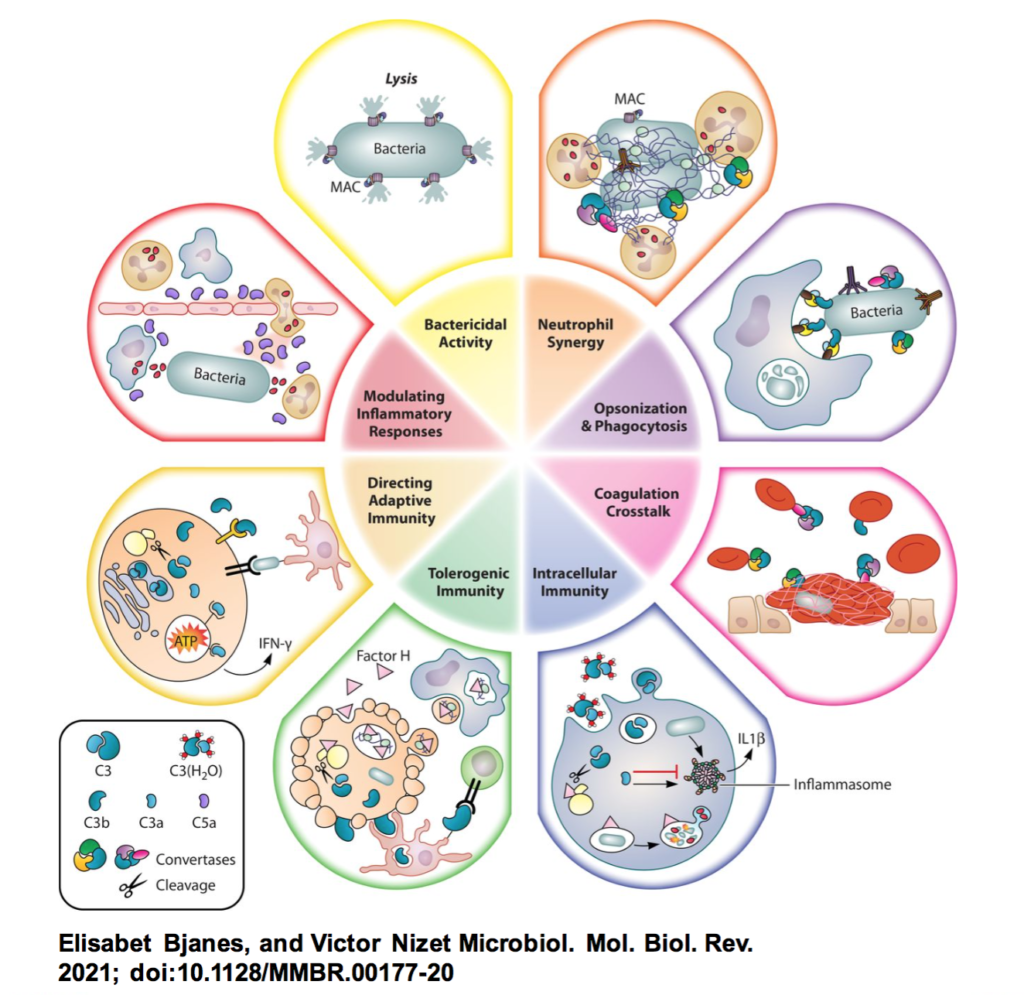
Review: More than a Pore: Nonlytic Antimicrobial Functions of Complement and Bacterial Strategies for Evasion – Elisabet Bjanes and Victor Nizet – Microbiology and Molecular Biology Reviews
*This review covers lytic and nonlytic roles of complement in the context of antimicrobial immunity and detail bacterial evasion mechanisms with a particular emphasis on clinically relevant Gram-positive pathogens.
Bacterial defenses against a natural antibiotic promote collateral resilience to clinical antibiotics – Lucas A. Meirelles – PlosOne
*Study on how natural toxins can promote tolerance and resistance to clinical antibiotics.
Review: Ancient Antibiotics, Ancient Resistance – Nicholas Waglechner – EcoSalPlus
Metabolism
Metabolic fitness landscapes predict the evolution of antibiotic resistance – Fernanda Pinheiro – Nature Ecology & Evolution
*Pinheiro et al. developed a metabolic model of antibiotic resistance that predicts the dosage-response of common resistance mutants and prevalent resistance mechanisms depending on drug levels and nutrient conditions. These predictions are confirmed by empirical growth inhibition curves and genomic data of Escherichia coli populations exposed to streptomycin challenge.
News & Views: Metabolic models predict evolutionary dynamics – Matthew Scott – Nature Ecology & Evolution
Horizontal gene transfer
Adaptive evolution of hybrid bacteria by horizontal gene transfer – Jeffrey J. Power – PNAS
A mobile genetic element increases bacterial host fitness by manipulating development – Joshua M Jones – eLife
Plasmids facilitate pathogenicity, not cooperation, in bacteria – Anna E. Dewar – ResearchSquare
Piggybacking on niche-adaptation improves the maintenance of multidrug resistance plasmids – Julia Kloos – Molecular Biology and Evolution
*The findings represent a new solution to the “plasmid paradox” and may contribute to an increased understanding of successful associations between specific E. coli lineages and MDR plasmids.
Antimicrobial peptides
Review: Chemically modified and conjugated antimicrobial peptides against superbugs – Wenyi Li – Chemical Society Reviews
Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations – Payel Das – Nature Biomedical Engineering
Novel antibiotics
Looking for Solutions to the Pitfalls of Developing Novel Antibacterials in an Economically Challenging System – Gilles Courtemanche – Microbiology Research
Enhancement of antibiotic production by co-cultivation of two antibiotic producing marine Vibrionaceae strains – Yannick Buijs – FEMS Microbiology Ecology
Low-cost drug discovery with engineered E. coli reveals an anti-mycobacterial activity of benazepril – Nadine Bongaerts – bioRxiv
Global health
Community-based antibiotic access and use in six low-income and middle-income countries: a mixed-method approach – Nga T T Do – Lancet Glob Health
Antibiotic resistance: Global health crisis and metagenomics – Shailendra Yadav and Atya Kapley – Biotechnology Reports
Human-associated microbiome
Human-associated microbiota suppress invading bacteria even under disruption by antibiotics – Andrew D. Letten – The ISME Journal
*Letten et al. found strong evidence of community-level suppression of the resistant strain in the absence of antibiotics and, despite large changes in community composition and abundance following rifampicin exposure, suppression of the invading resistant strain was maintained in both antibiotic treatments.
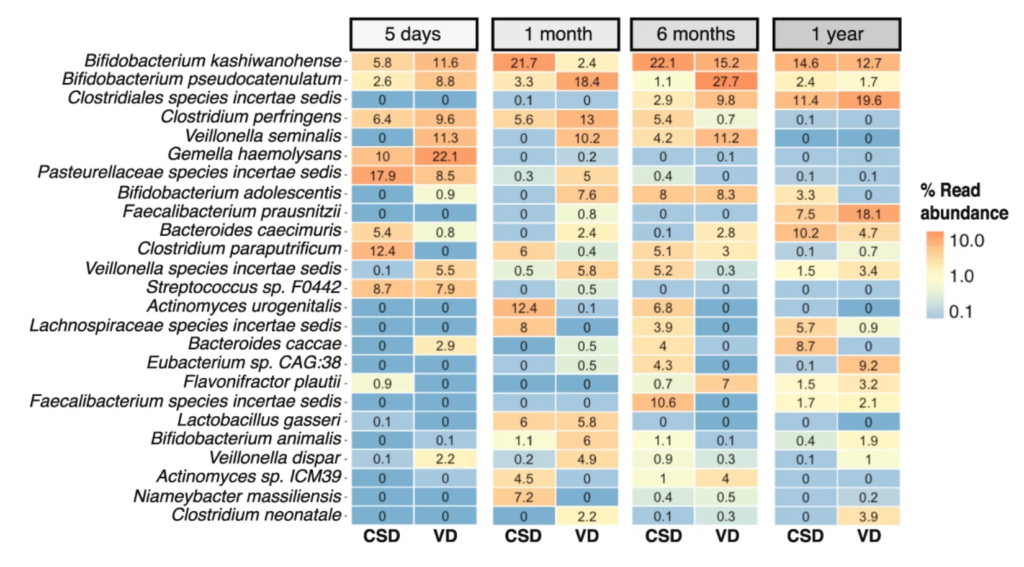
Persistence of birth mode-dependent effects on gut microbiome composition, immune system stimulation and antimicrobial resistance during the first year of life – Susheel Bhanu Busi, Laura de Nies – ISME Communications
*The study follows the babies from 5 days after birth through one year of age. Caesarean-section babies had an over-abundance of “synthetic” antibiotic resistance genes.
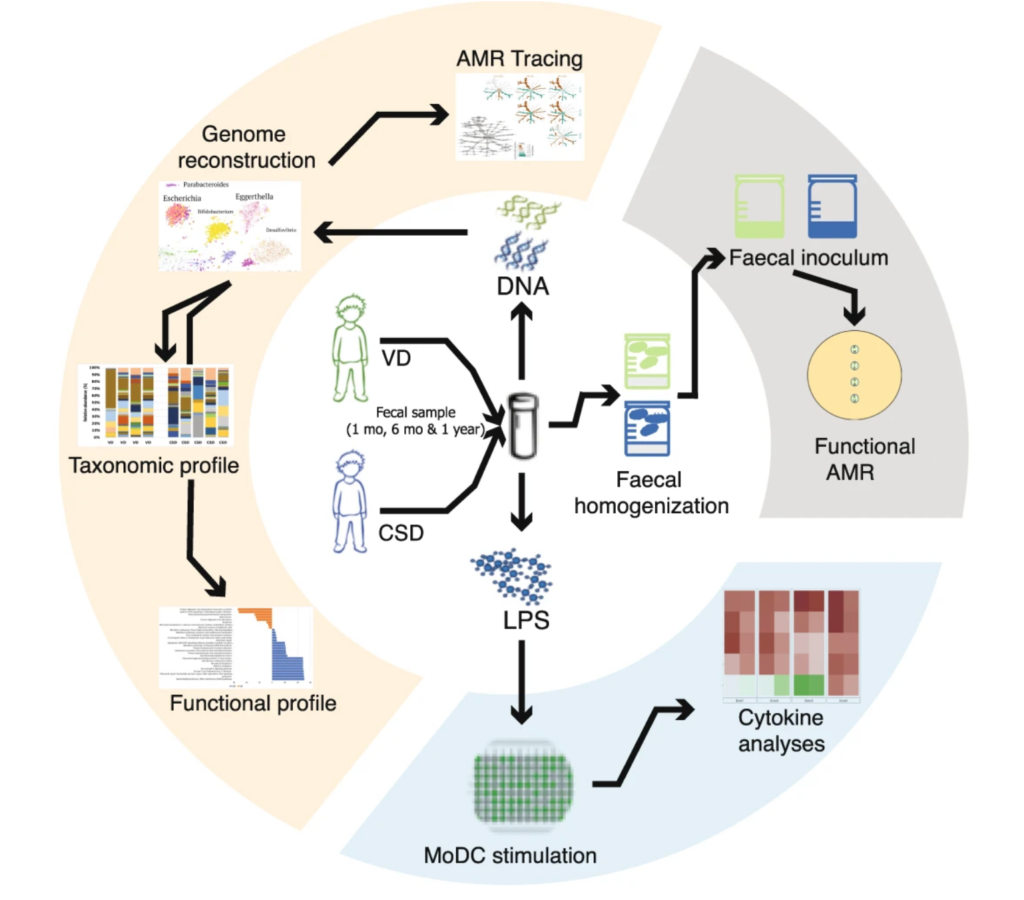
Review: Association between antibiotics and gut microbiome dysbiosis in children: systematic review and meta-analysis – Lucy McDonnell – Gut Microbes
Review: Early life antibiotics and childhood gastrointestinal disorders: a systematic review – Kim Kamphorst – BMJ Paediatrics Open
Whole-genome analysis of probiotic product isolates reveals the presence of genes related to antimicrobial resistance, virulence factors, and toxic metabolites, posing potential health risks – Ying Wang – BMC Genomics
Clinical environment
Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries – Kirsty Sands – Nature Microbiology
Pervasive transmission of a carbapenem resistance plasmid in the gut microbiota of hospitalized patients – Ricardo León-Sampedro – Nature Microbiology
Investigating potential transmission of antimicrobial resistance in an open-plan hospital ward: a cross-sectional metagenomic study of resistome dispersion in a lower middle-income setting – Anushia Ashokan– Antimicrobial Resistance & Infection Control
Economic Evaluation: Economic Burden of Urinary Tract Infections From Antibiotic-Resistant Escherichia coli Among Hospitalized Adult Patients in Lebanon: A Prospective Cohort Study – Katia Iskandar – Value in Health Regional Issues
Apparent nosocomial adaptation of Enterococcus faecalis predates the modern hospital era – Anna K. Pöntinen – Nature Communications
Antibiotic intervention redisposes bacterial interspecific interacting dynamics in competitive environments – Jing Li – Environmental Microbiology
The evolutionary trade‐offs in phage‐resistant Klebsiella pneumoniae entail cross‐phage sensitization and loss of multidrug resistance – Grazyna Majkowska‐Skrobek – Environmental Microbiology
Resistance to aztreonam in combination with non-β-lactam β-lactamase inhibitors due to the layering of mechanisms in Escherichia coli identified following mixed culture selection – Ching Hei Phoebe Cheung – bioRxiv
Multiple classes of bactericidal antibiotics cause DNA double strand breaks in Staphylococcus aureus – Rebecca S Clarke and Andrew M Edwards – bioRxiv
Comparing treatment strategies to reduce antibiotic resistance in an in vitro epidemiological setting – Daniel C. Angst – PNAS
Discontinuing β-lactam treatment after 3 days for patients with community-acquired pneumonia in non-critical care wards (PTC): a double-blind, randomised, placebo-controlled, non-inferiority trial – Aurélien Dinh – The Lancet
Animal microbiome
Differences in the gut microbiomes of dogs and wolves: roles of antibiotics and starch – Yuting Liu – BMC Veterinary Research
Transmission of Similar Mcr-1 Carrying Plasmids among Different Escherichia coli Lineages Isolated from Livestock and the Farmer – Joaquim Viñes – Antibiotics
*This work highlights the role of livestock as a reservoir of pathotypes with zoonotic potential.
Soil and plant microbiome
Vertical distribution of antibiotic resistance genes in an urban green facade – Shu-Yi-Dan Zhou – Environment International
*Study suggests that microorganisms may move from soil to plant, potentially through an endophytic mechanism and thus, there is a clear potential for movement of ARGs and human pathogens from the outdoor environment.
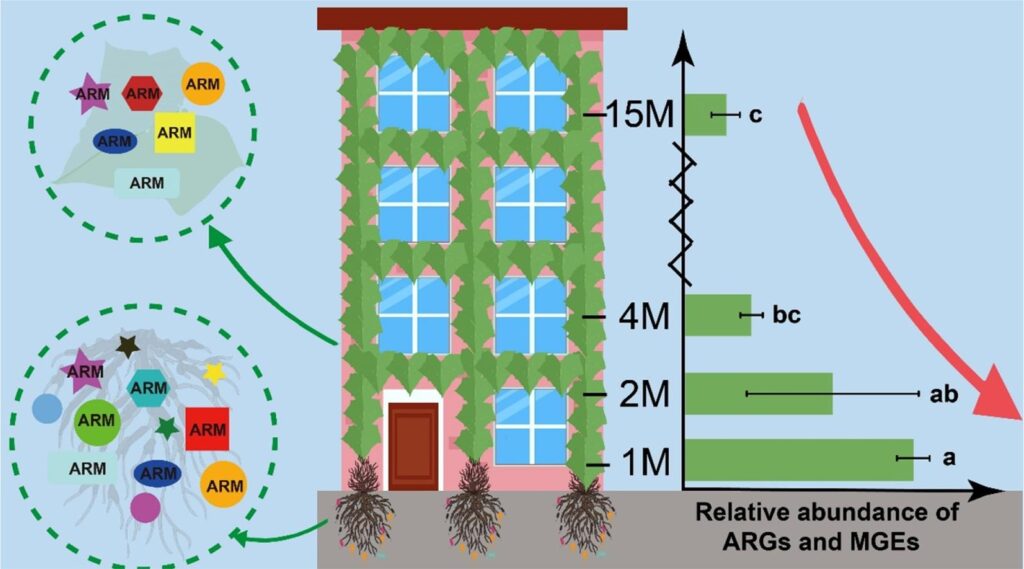
Targeted metagenomics reveals inferior resilience of farm soil resistome compared to soil microbiome after manure application – Gonçalo Macedo – Science of The Total Environment
Water environment
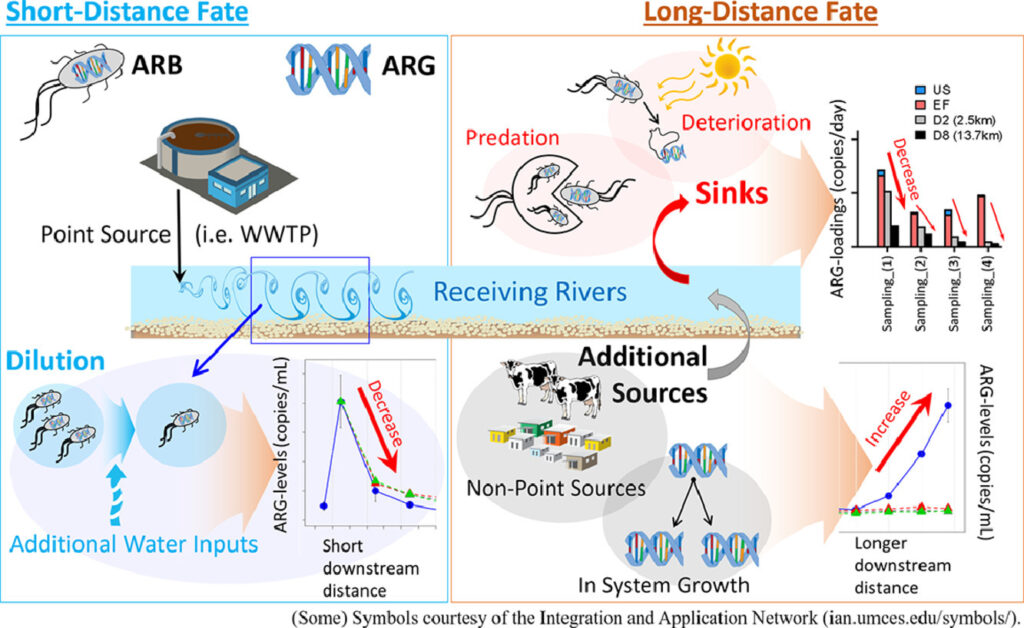
Unraveling the riverine antibiotic resistome: the downstream fate of anthropogenic inputs – Jangwoo Lee – Water Research
*Great work on drivers of riverine resistomes, pinpointing monitoring targets to identify where human sources and exposures are likely to be most acute. The authors observed the increase of certain gene markers downstream, without obvious sources of anthropogenic contamination.
Review: A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments – Dongsheng Zheng – Science of The Total Environment
Responses of bacterial and bacteriophage communities to long-term exposure to antimicrobial agents in wastewater treatment systems – Bingbing Du – Journal of Hazardous Materials
A Peek into the Plasmidome of Global Sewage – Philipp Kirstahler – bioRxiv
Biotransformation of lincomycin and fluoroquinolone antibiotics by the ammonia oxidizers AOA, AOB and comammox: A comparison of removal, pathways, and mechanisms – Li-Jun Zhou – Water Research
Selective enrichment of comammox from activated sludge using antibiotics – Yulin Wang – Water Research
Viruses and phages
Systematic exploration of Escherichia coli phage-host interactions with the BASEL phage collection – Enea Maffei – bioRxiv
*The BASEL (BActeriophage SElection for your Laboratory) is a collection of representative E. coli phages from all major families with their systematic phenotypic and genomic characterization.
The evolutionary trade‐offs in phage‐resistant Klebsiella pneumoniae entail cross‐phage sensitization and loss of multidrug resistance – Grazyna Majkowska‐Skrobek – Environmental Microbiology
Rapid evolutionary turnover of mobile genetic elements drives microbial resistance to viruses – Fatima Aysha Hussain – bioRxiv
Bioinformatics
The extent of carbapenemase-encoding genes in public genome sequences – Ingmar Janse – PeerJ
Origin Sample Prediction and Spatial Modeling of Antimicrobial Resistance in Metagenomic Sequencing Data – Maya Zhelyazkova – Frontiers in Genetics
Machine-learning prediction of resistance to sub-inhibitory antimicrobial concentrations from Escherichia coli genomes – Sam Benkwitz-Bedford – bioRxiv
Protocols
Podcast
February 2021 Pod: Global Change – Bengtsson-Palme Lab – Microbiology Lab Pod
Ep 26. Mirko Ancillotti & bioethics. Resistance to antibiotics vs. vaccines. – Uppsala Antibiotic Center – The AMR Studio
Ep 84. The Dangers of Antibiotic Resistance – Sean Allen and Nathan Allen – Petri Dish
Webinars
University of Nottingham Sutton Bonington AMR Webinar Series 2021
UAC Seminar: The visual culture of antimicrobial resistance by Rachel Irwin – Uppsala Antibiotic Center – April 9
Fun
Dr Dirt – a simulation game from Hoskisson Lab and Game Doctor team on antibiotic discovery
And the Microbes – a comic by Edward Ross and Jamie Hall: Join Luna and her sulky brother Simon on a journey into the weird and wonderful world of microbes.
