prepared by Marcus Wenne
In December’s AMR digest we ask a wide range of questions. Why do antibiotics exist? What effect do they have on our own microbiota? Can they increase the colonizing capability of plasmid carrying bacteria? And what methods can we use to measure resistance against them? This is a small sample of the wide range of topics in this month’s digest.
Antibiotic research and development
Review: Prospects for Antibacterial Discovery and Development – Thomas M Privalsky – Journal of the American Chemical Society
Why Do Antibiotics Exist? – Fabrizio Spagnolo – mBio
Global
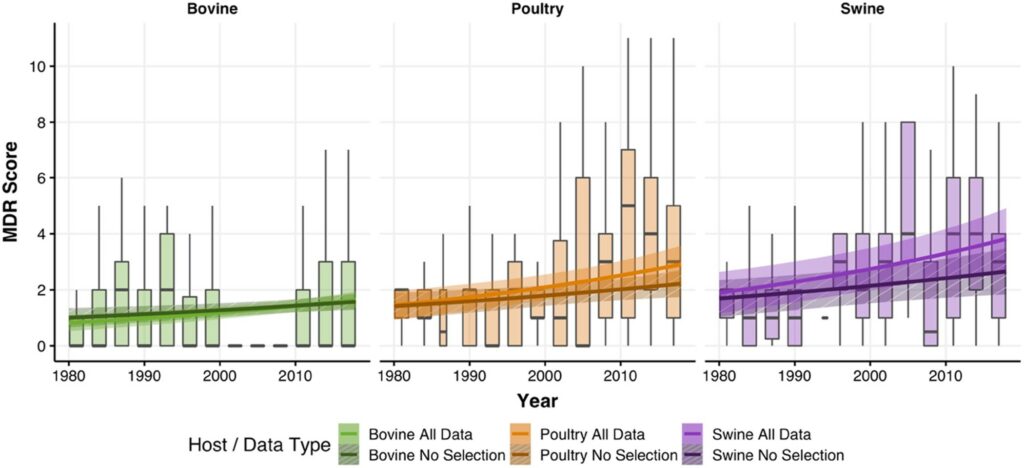
Increase in antimicrobial resistance in Escherichia coli in food animals between 1980 and 2018 assessed using genomes from public databases – João Pires – Journal of Antimicrobial Chemotherapy
*This study utilized public databases to infer the level of multi drug resistance in E .coli on a global scale from 1980 to 2018. The authors detected an increase in the number of antibiotics E. coli tended to carry resistance against (MDR score) by 1.6 times during this time frame.
Evolution of ColE1-like plasmids across γ-Proteobacteria: From bacteriocin production to antimicrobial resistance – Manuel Ares-Arroyo – Plos Genetics
Global protein responses of multi-drug resistant plasmid containing Escherichia coli to ampicillin, cefotaxime, imipenem and ciprofloxacin – Anatte Margalit – Journal of Global Antimicrobial Resistance
Towards the biogeography of prokaryotic genes – Luis Pedro Coelho – Nature
Clinical
Correlation between phenotypic virulence traits and antibiotic resistance in Pseudomonas aeruginosa clinical isolates – Osama Nassar – Microbial Pathogenesis
Antimicrobial resistance in commensal opportunistic pathogens isolated from non-sterile sites can be an effective proxy for surveillance in bloodstream infections – Karina-Doris Vihta – Scientific Reports
Identification of antibiotic collateral sensitivity and resistance interactions in population surveillance data – Laura B Zwep – JAC-Antimicrobial Resistance
Human Microbiome
Combinatorial, additive and dose-dependent drug–microbiome associations – Sofia K Forslund – Nature
*This study used a multi-omics approach to investigate drug-microbiome interactions in patients suffering from cardiometabolic disease. The authors found that multiple antibiotic exposures had cumulative effects on the patients microbiome. Leading to reduced richness and increased resistance, which are hallmarks of microbiota in patients suffering from obesity, insulin resistance and low-grade inflammation.
Influence of timing of maternal antibiotic administration during caesarean section on infant microbial colonisation: a randomised controlled trial – Thomas Dierikx – Gut Microbiota
Agriculture
Implications of the use of organic fertilizers for antibiotic resistance gene distribution in agricultural soils and fresh food products. A plot-scale study – Claudia Sanz – Science of The Total Environment
AMR detection methods
Review: Progressing Antimicrobial Resistance Sensing Technologies across Human, Animal, and Environmental Health Domains – Kira J Fitzpatrick – Journal of the American Chemical Society
Integrating Scalable Genome Sequencing Into Microbiology Laboratories for Routine Antimicrobial Resistance Surveillance – Mihir Kekre – Clinical Infectious Diseases
Review: Engineered CRISPR-Cas systems for the detection and control of antibiotic-resistant infections – Yuye Wu – Journal of Nanobiotechnology
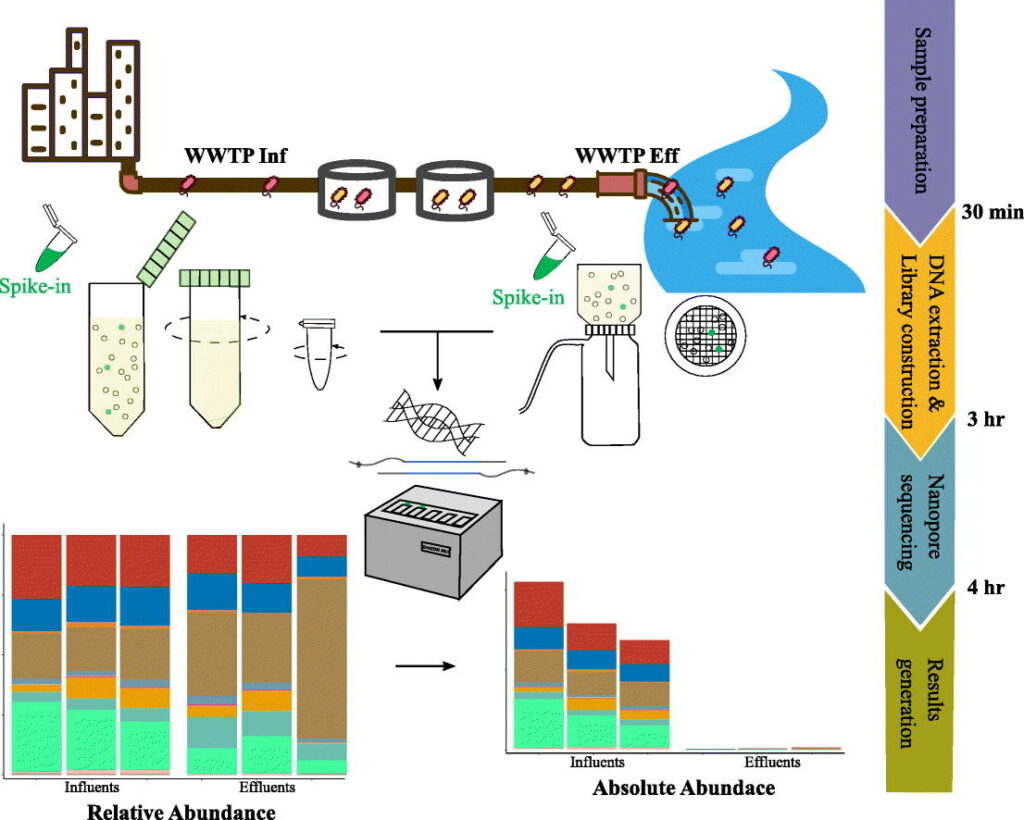
Rapid absolute quantification of pathogens and ARGs by nanopore sequencing – Yu Yang – The Science of the Total Environment
*Sequencing has become cheaper, faster and more accessible these last few decades. With the use of third generation sequencing these authors test a method where they can quantify the absolute abundance of both pathogens and resistance genes in 4 hours using third generation nanopore sequencing. Taking advantage of the nanopore technologies long reads, the authors were able to link resistance genes to individual taxa.
Sewage treatment
The physiological and ecological properties of bacterial persisters discovered from municipal sewage sludge and the potential risk – Xiang Liu – Environmental Research
Water environment
Metagenomic profiles of the resistome in subtropical estuaries: Co-occurrence patterns, indicative genes, and driving factors – Lei Zhou – Science of The Total Environment
Assessment on impact of sewage in coastal pollution and distribution of fecal pathogenic bacteria with reference to antibiotic resistance in the coastal area of Cape Comorin, India – Nanthini S Victoria – Marine Pollution Bulletin
Animal experiment
Colonization of gut microbiota by plasmid-carrying bacteria is facilitated by evolutionary adaptation to antibiotic treatment – Peng Zhang – The ISME Journal
Courses
Webinars
Future conferences
Podcasts