AMR JUNE, 2022
prepared by Víctor Hugo Jarquín-Díaz
For those in the northern hemisphere… Summer is here ⛱️! With it, the EMBARK team brings a refreshing AMR digest with many sunshiny papers. Put your 🕶️ on and swim deep into many works that we have compiled for you: AMR in soil, water, clinical or livestock settings, antibiotic resistance gene ecology and evolution and more … for those in the southern hemisphere, we also wish you a happy reading despite the cold winter ^^
Global
*Addressing a future pandemic: how can non-biological complex drugs prepare us for antimicrobial resistance threats? – Blackman, L. D., Sutherland, T. D., De Barro, P. J., Thissen, H., & Locock, K. E. – Materials Horizons
Lewis D. Blackman et al. review the different ways in which bacteria develop resistance against antibiotics and alternative agents that could be employed. They focused on non-biological complex drugs (NBCDs) as the next generation antimicrobial agents. They outline the advancements in antimicrobial polymer materials, carbon nanomaterials, and inorganic nanomaterials and highlight the remaining challenges for their clinical translation.
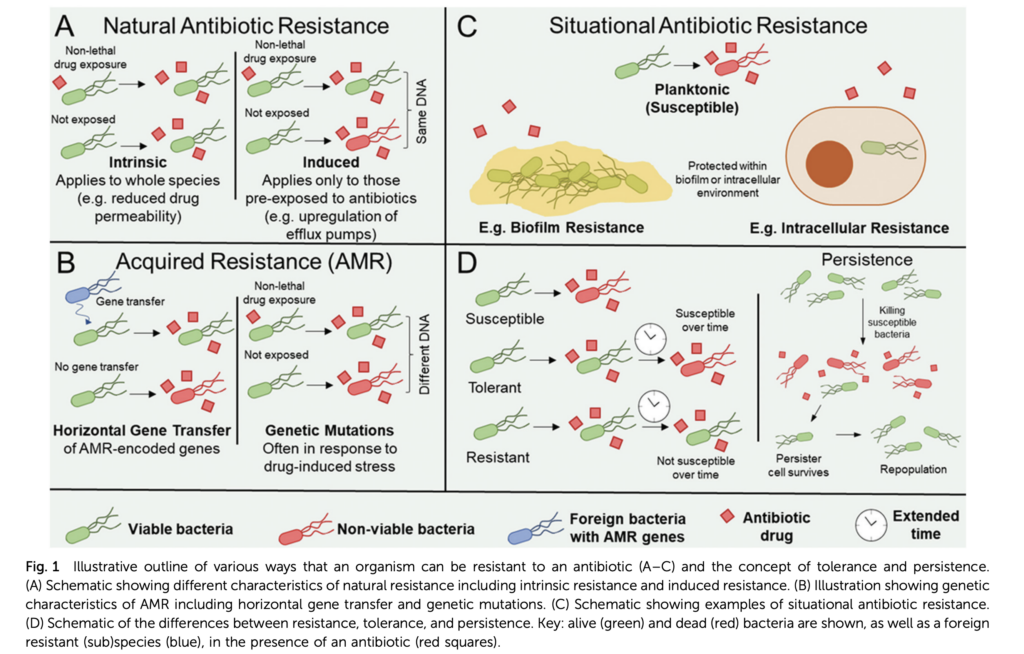
*Environmental Dimensions of “One Health” to Combat Antimicrobial Resistance: Essential Research Needs – Jin, L., Pruden, A., Boehm, A. B., Alvarez, P. J., Raskin, L., Kohn, T., & Li, X. – Integrating Environmental Science & Technology
In this viewpoint article, Jin et al. critically address the lack of relevance and impact on health outcomes in environmental AMR research. They invite to “better define impactful contributions from the environmental dimension of AMR as part of a broader “One Health” vision” by implementing and developing models in line with an anthropocentric context of bacterial transfer within the interface between environment and humans. The authors promote multidisciplinary cross-national participation and coordination to establish analytical approaches to interpret ARG in the environment comprehensively. Approaches to identify responsible agents or conditions that select for AMR evolution and fit-for-purpose treatment technologies for mitigating high-risk ARGs and ARB at crucial sources.
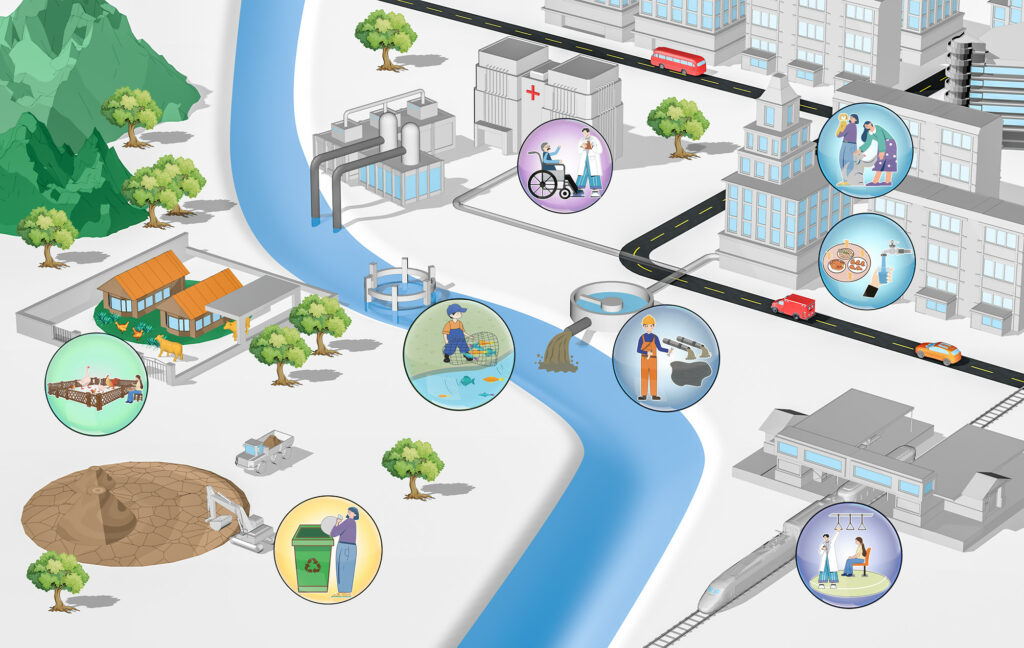
*A bottom-up view of antimicrobial resistance transmission in developing countries – Ikhimiukor, O.O., Odih, E.E., Donado-Godoy, P. and Iruka N. Okeke – Nature Microbiology
This review from Odion O. Ikhimiukor et al. focuses on AMR transmission in low- and middle-income countries, emphasizing high-risk transmission points such as urban settings and food-animal handling. The authors describe the integration of top-down and bottom-up strategies as AMR-containment approaches. They suggest that technological innovations are required to control AMR in low- and middle-income settings.
Editorial: Antimicrobial resistance – da Silva Dantas, A. – Molecular Microbiology
Global burden of antimicrobial resistance: essential pieces of a global puzzle – Charani, E., McKee, M., Balasegaram, M., Mendelson, M., Singh, S., & Holmes, A. – The Lancet
Soil antimicrobial resistance
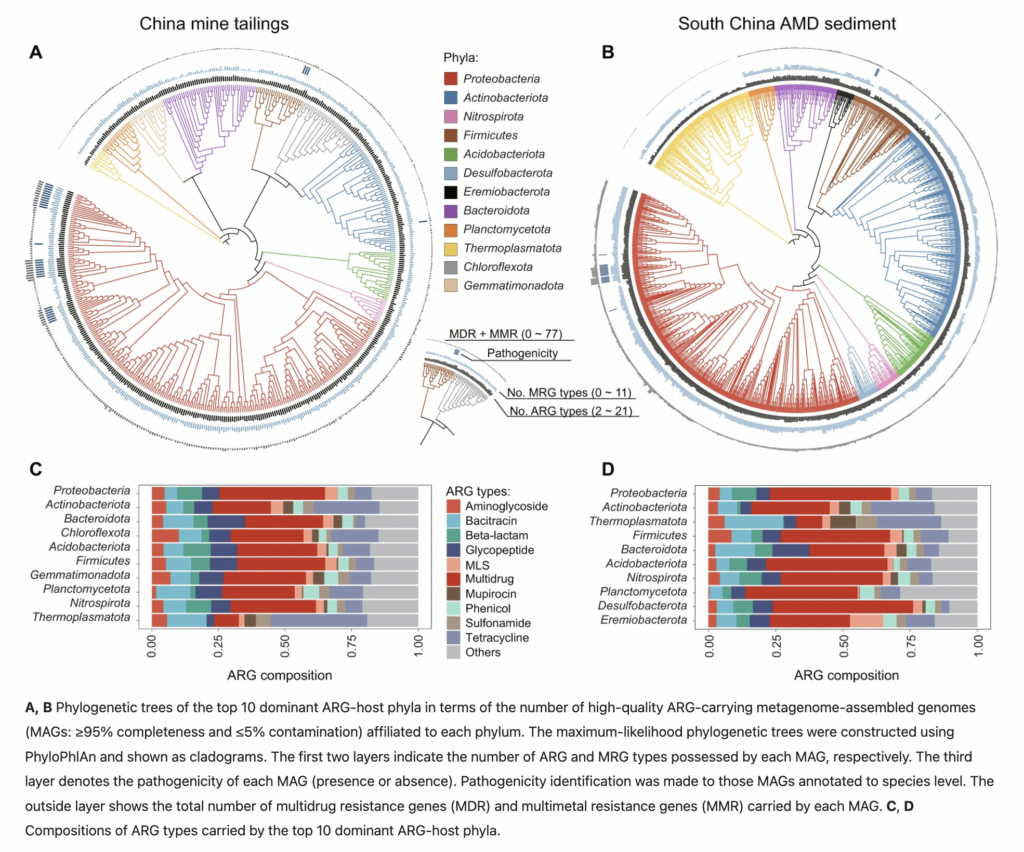
*Globally distributed mining-impacted environments are underexplored hotspots of multidrug resistance genes – Yi, X., Liang, J. L., Su, J. Q., Jia, P., Lu, J. L., Zheng, J. & Zhu, Y. G. – The ISME Journal
Yi et al. focused their work on giving further insight into antimicrobial resistance in mining-impacted environments. They suggest developing constant monitoring strategies as the mining sites represent an underexplored hotspot for multidrug resistance genes. Overall, they described around 54 high-quality ARG-carrying MAGs from the phylum Proteobacteria, Acidobacteria, Actinobacteria, Bacteroidota, Firmicutes, Nitrospira, Planctomycetes and Thermoplasmatota and confirmed high mobility of ARGs, mainly trough transposons and plasmid.
Deciphering environmental resistome and mobilome risks on the stone monument: A reservoir of antimicrobial resistance genes – He, J., Zhang, N., Shen, X., Muhammad, A., & Shao, Y. – Science of The Total Environment
Plant cultivar determined bacterial community and potential risk of antibiotic resistance gene spread in the phyllosphere – Fan, X., Su, J., Zhou, S., An, X., & Li, H. – Journal of Environmental Sciences
β-lactamase genes transmission influenced by tetracycline, sulfonamide and β-lactams antibiotics contamination in the on-site farm soil – Qi, Z., Le, Z., Han, F., Qi, Y., & Liu, R. – Ecotoxicology and Environmental Safety
Effect of pesticides on nitrification activity and its interaction with chemical fertilizer and manure in long-term paddy soils – Mukhles, M. B., Rahman, M. M., Rana, M. R., Huda, N., Ferdous, J., Rahman, F., & Biswas, S. K. – Chemosphere
Insights into microbial contamination in multi-type manure-amended soils: the profile of human bacterial pathogens, virulence factor genes and antibiotic resistance genes – Zhu, L., Lian, Y., Lin, D., Huang, D., Yao, Y., Ju, F., & Wang, M. – Journal of Hazardous Materials
Water environment and waste water treatment
Exploring the microbiome, antibiotic resistance genes, mobile genetic element, and potential resistant pathogens in municipal wastewater treatment plants in Brazil – Leroy-Freitas, D., Machado, E. C., Torres-Franco, A. F., Dias, M. F., Leal, C. D., & Araújo, J. C . – Science of The Total Environment
Genomic Analysis of Carbapenem-Resistant Comamonas in Water Matrices: Implications for Public Health and Wastewater Treatments – Hem, S., Wyrsch, E. R., Drigo, B., Baker, D. J., Charles, I. G., Donner, E., & Djordjevic, S. P. – Applied and Environmental Microbiology
Metagenomic assembly and binning analyses the prevalence and spread of antibiotic resistome in water and fish gut microbiomes along an environmental gradient – Guan, Y., Xue, X., Jia, J., Li, X., Xing, H., & Wang, Z. – Journal of Environmental Management
Wastewater plastisphere enhances antibiotic resistant elements, bacterial pathogens, and toxicological impacts in the environment – Junaid, M., Liu, S., Liao, H., Liu, X., Wu, Y., & Wang, J. – Science of The Total Environment
Antimicrobial Resistance Monitoring of Water Environments: A Framework for Standardized Methods and Quality Control – Liguori, K., Keenum, I., Davis, B. C., Calarco, J., Milligan, E., Harwood, V. J., & Pruden, A. – Environmental Science & Technology
Air environment and airborne ARGs
A review of the emergence of antibiotic resistance in bioaerosols and its monitoring methods – Lee, G., & Yoo, K. – Reviews in Environmental Science and Bio/Technology
Effects of antibiotics consumption on the behavior of airborne antibiotic resistance genes in chicken farms – Song, L., Jiang, G., Wang, C., Ma, J., & Chen, H. – Journal of Hazardous Materials
Phages and antimicrobial resistance
Antibiotic Exposure Leads to Reduced Phage Susceptibility in Vancomycin Intermediate Staphylococcus aureus (VISA) – McCallin, S., Menzi, C., Lassen, S., Daraspe, J., Oechslin, F., & Moreillon, P. – Antimicrobial Agents and Chemotherapy
Isolation of a lytic bacteriophage against extensively drug‐resistant Acinetobacter baumannii infections and its dramatic effect in rat model of burn infection – Ghaznavi-Rad, E., Komijani, M., Moradabadi, A., Rezaei, M., & Shaykh-Baygloo, N. – Journal of Clinical Laboratory Analysis
Domestic animals, “Farm-to-fork” ARG transmission and wildlife
Multiresistant Enterobacteriaceae in yellow-legged gull chicks in their first weeks of life – Vittecoq, M., Brazier, L., Elguero, E., Bravo, I. G., Renaud, N., Manzano‐Marín, A., & Thomas, F. – Ecology and Evolution
Effect of antimicrobial administration on fecal microbiota of critically ill dogs: dynamics of antimicrobial resistance over time – Menard, J., Goggs, R., Mitchell, P., Yang, Y., Robbins, S., Franklin-Guild, R. J., & Goodman, L. B. – Animal Microbiome
Expressions of resistome is linked to the key functions and stability of active rumen microbiome – Ma, T., Zaheer, R., McAllister, T. A., Guo, W., Li, F., Tu, Y., & Guan, L. L. – Animal Microbiome
Role of Horizontal Gene Transfer in the Dissemination of Antimicrobial Resistance in Food Animal Production – Vinayamohan, P. G., Pellissery, A. J., & Venkitanarayanan, K. – Current Opinion in Food Science
High Throughput Screening of Antimicrobial Resistance Genes in Gram-Negative Seafood Bacteria – Delannoy, S., Hoffer, C., Youf, R., Dauvergne, E., Webb, H. E., Brauge, T., & Brisabois, A. – Microorganisms
Bacillus licheniformis–fermented products and enramycin differentially modulate microbiota and antibiotic resistome in the cecal digesta of broilers – Chen, Y. C., & Yu, Y. H. – Poultry Science
Metagenomic analysis of the gut microbiota in piglets either challenged or not with enterotoxigenic Escherichia coli reveals beneficial effects of probiotics on microbiome composition, resistome, digestive function and oxidative stress responses – Apiwatsiri P, Pupa P, Sirichokchatchawan W, Sawaswong V, Nimsamer P, Payungporn S, et al. – PLoS ONE
Antibiotic resistance in clinical settings
Metagenomic DNA sequencing for semi-quantitative pathogen detection from urine: a prospective, laboratory-based, proof-of-concept study – Janes, V. A., Matamoros, S., Munk, P., Clausen, P. T., Koekkoek, S. M., Koster, L. A., & Schultsz, C. – The Lancet Microbe
A resistome roadmap: from the human body to pristine environments – Maestre-Carballa, L., Navarro-López, V., & Martinez-Garcia, M. – Frontiers in Microbiology
Antibiotic-resistant organisms establish reservoirs in new hospital built environments and are related to patient blood infection isolates – Sukhum, K. V., Newcomer, E. P., Cass, C., Wallace, M. A., Johnson, C., Fine, J., & Kwon, J. H. – Communications Medicine
Economic burden of antibiotic-not-susceptible isolates in uncomplicated urinary tract infection: Analysis of a US integrated delivery network database – Shafrin, J., Marijam, A., Joshi, A.V. – Antimicrobial Resistance & Infection Control
Fusion plasmid carrying the colistin resistance gene mcr of Escherichia coli isolated from healthy residents – Hoa, H. T. T., Higashi, A., Yamaguchi, T., Kawahara, R., Calvopina, M., Bastidas-Caldés, A., & Yamamoto, Y. – Journal of Global Antimicrobial Resistance
An Optogenetic Toolkit for Light-Inducible Antibiotic Resistance – Sheets, M. B., & Dunlop, M. J – bioRxiv
Attributable mortality of vancomycin resistance in ampicillin-resistant Enterococcus faecium bacteremia in Denmark and the Netherlands: A matched cohort study – Rottier, W. C., Pinholt, M., van der Bij, A. K., Arpi, M., Blank, S. N., Nabuurs-Franssen, M. H., & Danish Collaborative Bacteraemia Network (DACOBAN) – Infection Control & Hospital Epidemiology
ARG ecology and evolution
GR13-type plasmids in Acinetobacter potentiate the accumulation and horizontal transfer of diverse accessory genes – Moran, R. A., Liu, H., Doughty, E. L., Hua, X., Cummins, E. A., Liveikis, T., McNally, A., Zhou, Z., van Schaik, W., Yu, Y. – Microbial Genomics
Exploring the Ecological Effects of Naturally Antibiotic-Insensitive Bifidobacteria in the Recovery of the Resilience of the Gut Microbiota during and after Antibiotic Treatment – Argentini, C., Mancabelli, L., Alessandri, G., Tarracchini, C., Barbetti, M., Carnevali, L., & Turroni, F. – Applied and Environmental Microbiology
Evolved resistance to a novel cationic peptide antibiotic requires high mutation supply – Santos-Lopez, A., Fritz, M. J., Lombardo, J. B., Burr, A. H., Heinrich, V. A., Marshall, C. W., & Cooper, V. S. – Evolution, Medicine, and Public Health
Localized pmrB hypermutation drives the evolution of colistin heteroresistance – Kapel, N., Caballero, J. D., & MacLean, R. C. – Cell Reports
Within-patient evolution of plasmid-mediated antimicrobial resistance – DelaFuente, J., Toribio-Celestino, L., Santos-Lopez, A., Leon-Sampedro, R., Alonso-del Valle, A. A., Costas, C., & San Millan, A. – bioRxiv
Effect of sulfamethazine on the horizontal transfer of plasmid-mediated antibiotic resistance genes and its mechanism of action – Yan, X., Liu, W., Wen, S., Wang, L., Zhu, L., Wang, J., & Wang, J. – Journal of Environmental Sciences
Phage-plasmids spread antibiotic resistance genes through infection and lysogenic conversion – Eugen Pfeifer, Remy Bonnin, Eduardo P. C. Rocha – bioRxiv
Bioinformatics
Datasets for benchmarking antimicrobial resistance genes in bacterial metagenomic and whole genome sequencing – Raphenya, A. R., Robertson, J., Jamin, C., de Oliveira Martins, L., Maguire, F., McArthur, A. G., & Hays, J. P. – Scientific Data
A genome-wide atlas of antibiotic susceptibility targets and pathways to tolerance – Leshchiner, D., Rosconi, F., Sundaresh, B., Rudmann, E., Ramirez, L. M. N., Nishimoto, A. T., & van Opijnen, T. – Nature Communications
Unraveling antimicrobial resistance using metabolomics – Kok, M., Maton, L., van der Peet, M., Hankemeier, T., & van Hasselt, J. C. – Drug Discovery Today
Webinar/Podcast
Join CIDRAP-ASP on June 21, 6:00 – 7:15 pm CDT for a webinar discussing the importance of effective, data-driven infection prevention strategies (IPC) and antibiotic stewardship (AS) initiatives. It has been shown that IPC and AS reduce drug-resistant infections and diminish the burden of antimicrobial resistance (AMR) within healthcare systems. More information here.
AMR Studio Podcast Ep 39: Vanessa Carter & patient advocacy. A stewardship game. Evolution of antibiotic tolerance – Uppsala Antibiotic Center