AMR APRIL, 2021
prepared by Saba Yasmin
Our April AMR digest features recent studies on antibiotics resistance in gut microbiota, prediction and quick diagnosis models as well as the role of birds and sewage water in AMR dissemination.
Join the SPRING AMR WEBINARS to discuss the latest AMR insights with us and our invited speakers. You can register HERE for the next webinar on May 19 with Heike Schmitt and Etienne Ruppé.
Global
Commentary: COVID-19: The therapeutic landscape – Catherine I. Paules and Anthony S. Fauci – Cell Press
Bioinformatics
ResistoXplorer: a web-based tool for visual, statistical, and exploratory data analysis of resistome data – Achal Dhariwal – NAR Genomics and Bioinformatics
*ResistoXplorer is a user-friendly tool that enables high-throughput analysis of common outputs generated from metagenomic resistome studies.
Forecasting the dissemination of antibiotic resistance genes across bacterial genomes – Mostafa M. H. Ellabaan – Nature Communications
*The authors predict the potential future dissemination of known ARGs by using statistical framework to identify putative horizontally transferred ARGs and the groups of bacteria that disseminate them. Analysis of ARG-surrounding sequences identify genes encoding putative mobilisation elements such as transposases and integrases that may be involved in gene transfer between genomes.
Rapid nanopore-based DNA sequencing protocol of antibiotic-resistant bacteria for use in surveillance and outbreak investigation – Fabienne Antunes Ferreira – Microbial Genomics
*This study developed and validated a complete analysis protocol for faster and more accurate surveillance and outbreak investigations of antibiotic-resistant microbes based on Oxford Nanopore Technologies (ONT) DNA whole-genome sequencing. The suggested protocol includes: (1) a 20 h sequencing run; (2) identification of the sequence type (ST); (3) de novo genome assembly; (4) polishing of the draft genomes; and (5) phylogenetic analysis based on SNPs.
Human associated microbiome
Signal vs. noise: how to analyze the microbiome and make progress on antimicrobial resistance – Jonathan L Golob and Krishna Rao – The Journal of Infectious Diseases
Dynamic Changes in Microbiome Composition Following Mare’s Milk Intake for Prevention of Collateral Antibiotic Effect – Almagul Kushugulova – Frontier in Cellular and Infectious Microbiology
*This study on the impact of mare’s milk consumption reveals it to be well tolerated in children concurrently receiving antibiotic treatment and suggests positive effects on recovery of the gut microbiome following such perturbation.
Resistome and microbial profiling of pediatric patient’s gut infected with multidrug-resistant diarrhoeagenic Enterobacteriaceae using next-generation sequencing; the first study from Pakistan – Ome Kalsoom Afridi – The Libyan Journal of Medicine
*The findings of the this study were an increased abundance of several pathogenic gram-negative bacteria namely E. coli, Enterobacter cloacae, and Salmonella enterica was observed in the gut microbiota of children infected with MDR bacterial infections than that of the healthy controls. This work indicates that children with MDR infections have reduced microbial diversity and enriched ARGs than healthy controls.
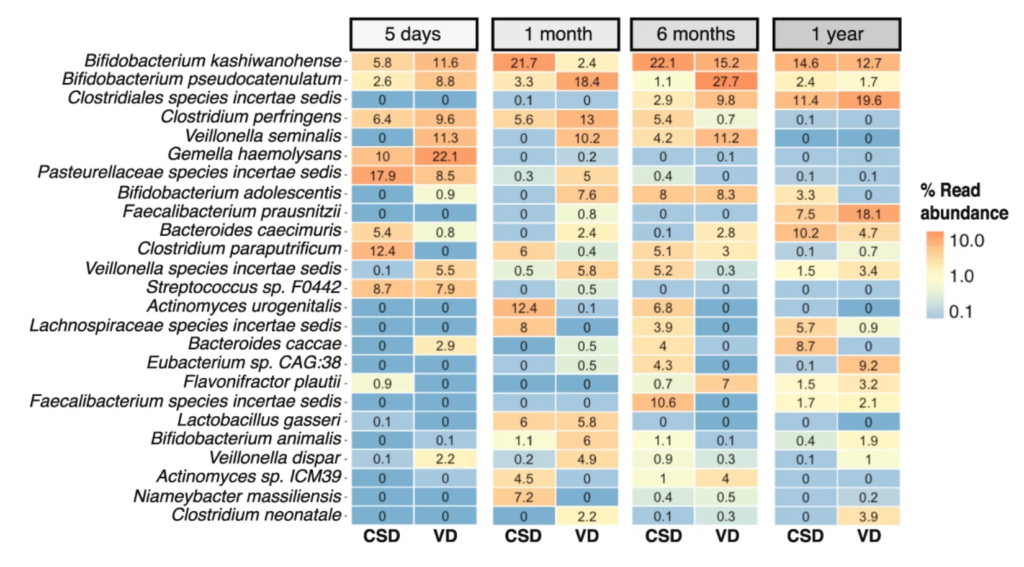
The infant gut resistome is associated with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition – Xuanji Li – Cell Host and Microbe
*Distribution of infant gut resistome is bimodal, mainly driven by E. coli. The infant gut resistome is significantly affected by environmental factors. Low maturity of microbiome associates with high ARG load. A similar asthma-associated gut bacterial composition associates with high ARG load.
Animal associated microbiome
Faecal microbiota and antibiotic resistance genes in migratory waterbirds with contrasting habitat use – Dayana Jarma – Science of the Total Environment
*The authors summarized that storks and gulls carry more antibiotic resistance genes than cranes and geese. Gulls carrying more pathogenic bacteria and resistance to last resort antibiotics.
Review: Epidemiology and Molecular Basis of Multidrug Resistance in Rhodococcus equi – Sonsiray Álvarez Narváez – Microbiology and Molecular Biology Reviews
*This review summarizes the factors that contributed to the development and spread of MDR R. equi, the molecular epidemiology of the emergence of MDR R. equi, the repercussions of MDR R. equi for veterinary and human medicine, and measures that might mitigate antimicrobial resistance at horse-breeding farms, such as alternative treatments to traditional antibiotics.
Deposition of resistant bacteria and resistome through FMT in germ-free piglets – Jing Sun – Letters in Applied Microbiology
Resistance to widely used disinfectants and heavy metals and cross resistance to antibiotics in Escherichia coli isolated from pigs, pork and pig carcass – Jiratchaya Puangseree – Food Control
*The authors concluded that MDR was predominant among the E. coli isolates (89%) from pig, pig carcass and pork. TCS MICs for E. coli in 2011–2014 were significantly higher than those in 2008–2010. MICs of biocides and heavy metals were weak correlated with antibiotic resistance. Exposure to biocides TCS, BKC and CHX contributes to antibiotic resistance.
Microbial genetics
Genetic determinants facilitating the evolution of resistance to carbapenem antibiotics – Peijun Ma – eLife
*In clinical isolates, the authors found that high-level transposon insertional mutagenesis plays an important role in contributing to high-level resistance frequencies in several major and emerging carbapenem-resistant lineages.
CRISPR-Cas is associated with fewer antibiotic resistance genes in bacterial pathogens – Elizabeth Pursey – BioRxiv
*The authors modelled the association between CRISPR-Cas and indicators of horizontal gene transfer and found that pathogens with a CRISPR-Cas system were less likely to carry antibiotic resistance genes than those lacking this defence system.
Environmental microbiology
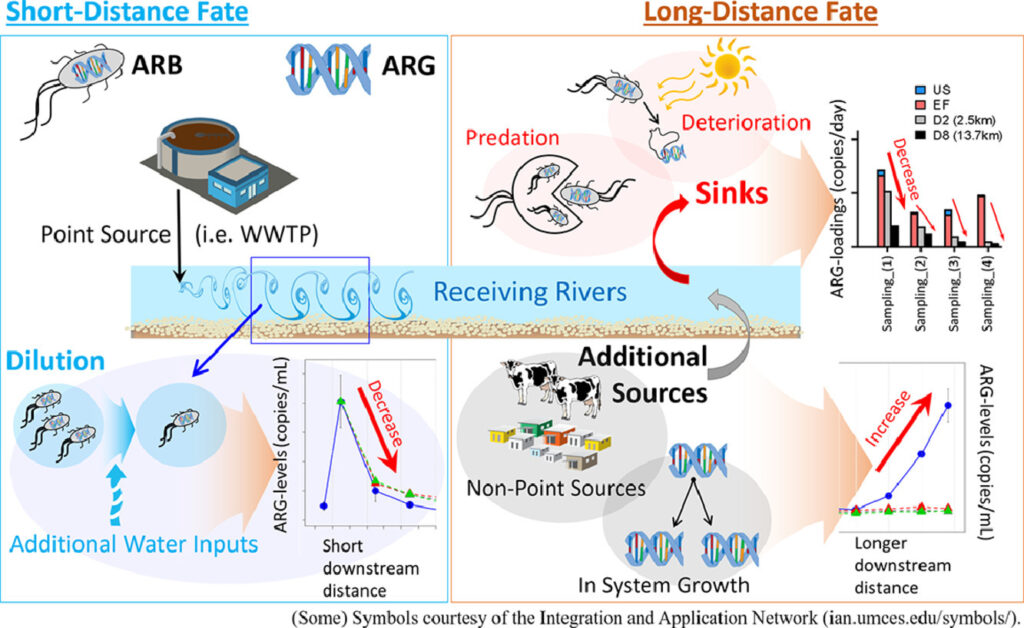
Review: Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges – Anh Q. Nguyen – Science of the Total Environment
*Wastewater treatment is a double-edged sword that can act as either a pathway for AMR spread or as a barrier to reduce the environmental release of anthropogenic AMR. This review highlights significant knowledge gaps including inconsistencies in ARG reporting units, lack of ARG/ARB monitoring surrogates, lack of a standardised protocol for determining ARG removal via wastewater treatments, and the inability to support appropriate risk assessment.
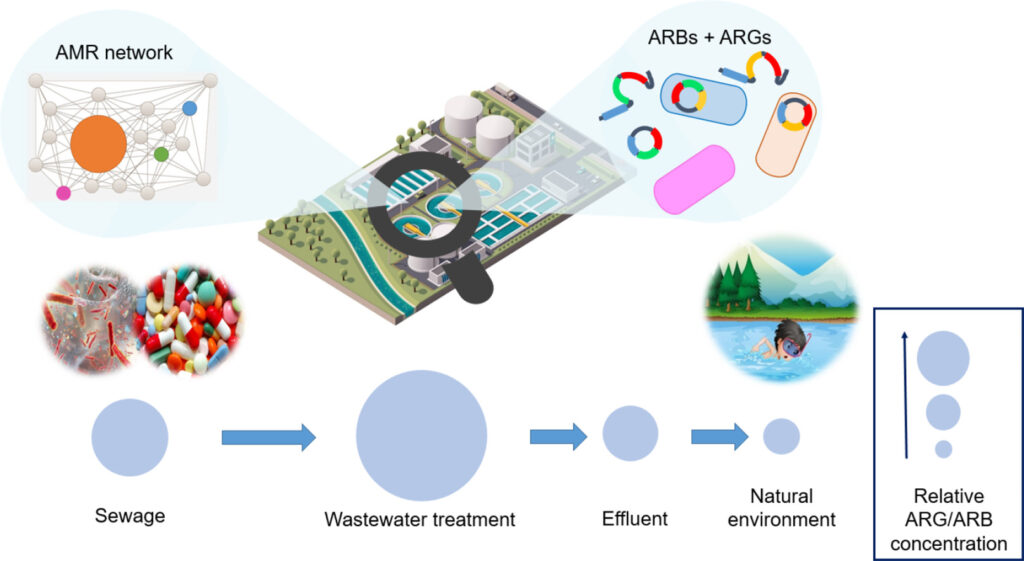
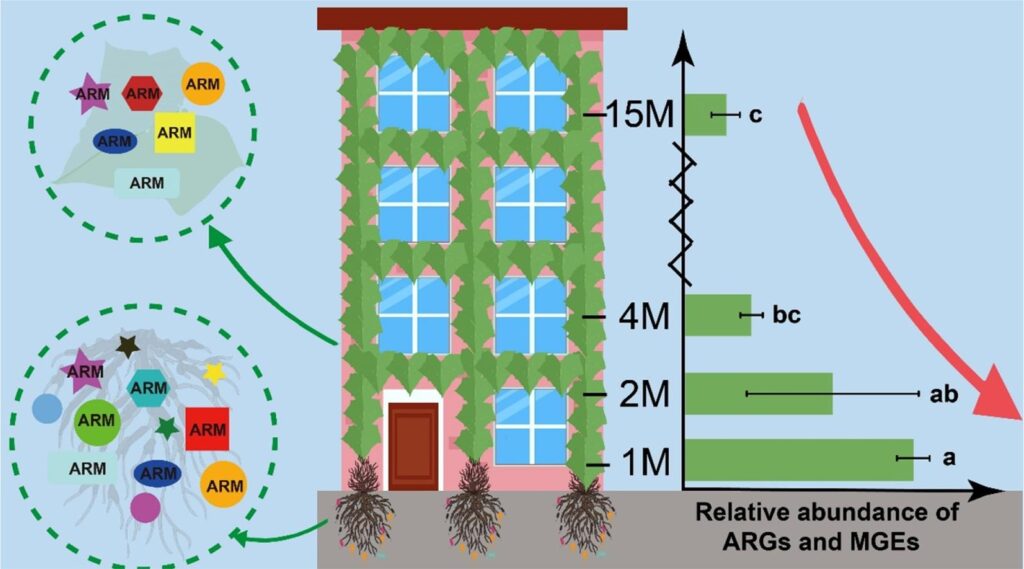
Spread of antibiotic resistance genes and microbiota in airborne particulate matter, dust, and human airways in the urban hospital – Zhen-Chao Zhou – Environment International
*The authors found that dust contained diverse antibiotic resistance genes (ARGs) and microbes. Airborne particulate matter contained high total relative abundance of ARGs. Microbes and ARGs in particulate matter posed high risks to patient airways.
Extended-Spectrum β-Lactamase and Carbapenemase Genes are Substantially and Sequentially Reduced during Conveyance and Treatment of Urban Sewage – Liguan Li – Environmental Science & Technology
Podcast
Episode X7 with Otto Cars, Eldar Shafir and Vanessa Carter: How do we change behaviors around antimicrobial resistance? – Uppsala Antibiotic Center – The AMR Studio
Webinar
The Natural History of Antibiotic Resistance – Gerry Wright – CARe, June 7


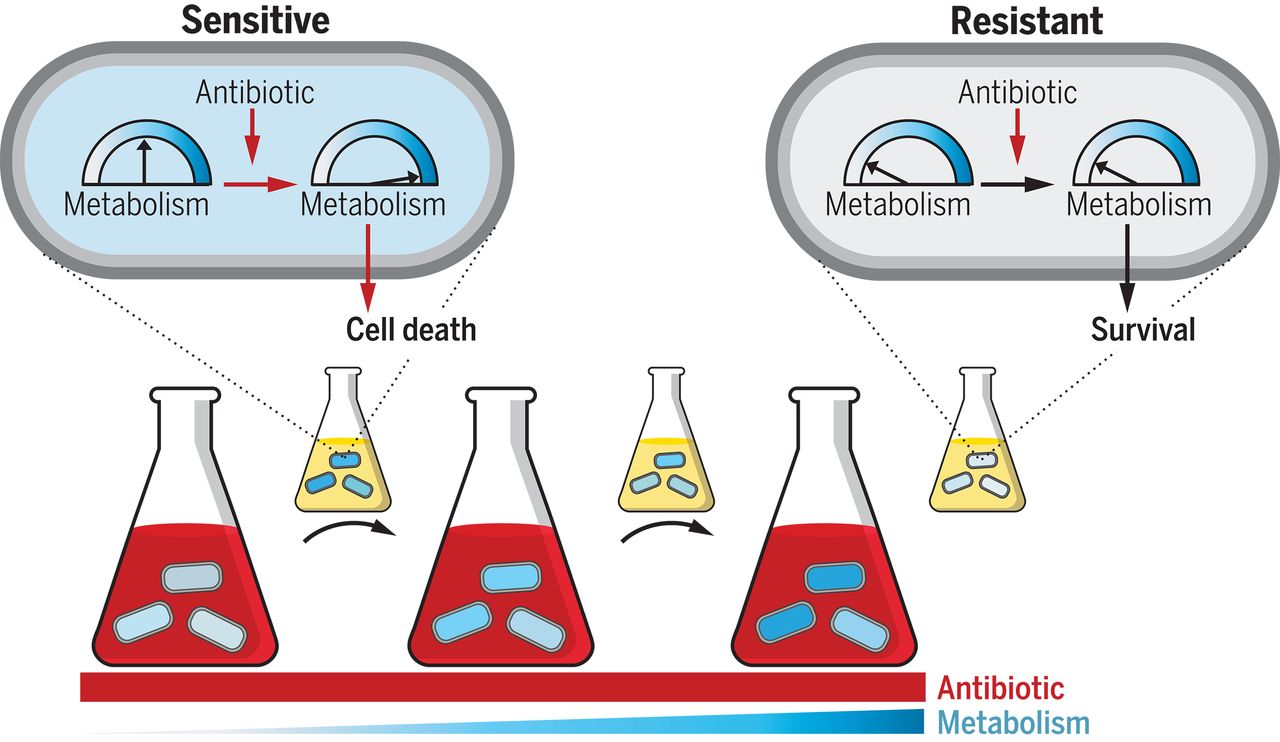 *Mutations targeting central carbon and energy metabolism arise in response to antibiotic treatment. These mutations confer resistance and are highly prevalent in clinical pathogens, expanding the known means by which pathogenic microbes can evolve resistance.
*Mutations targeting central carbon and energy metabolism arise in response to antibiotic treatment. These mutations confer resistance and are highly prevalent in clinical pathogens, expanding the known means by which pathogenic microbes can evolve resistance.